publications
publications by categories in reversed chronological order. generated by jekyll-scholar.
2025
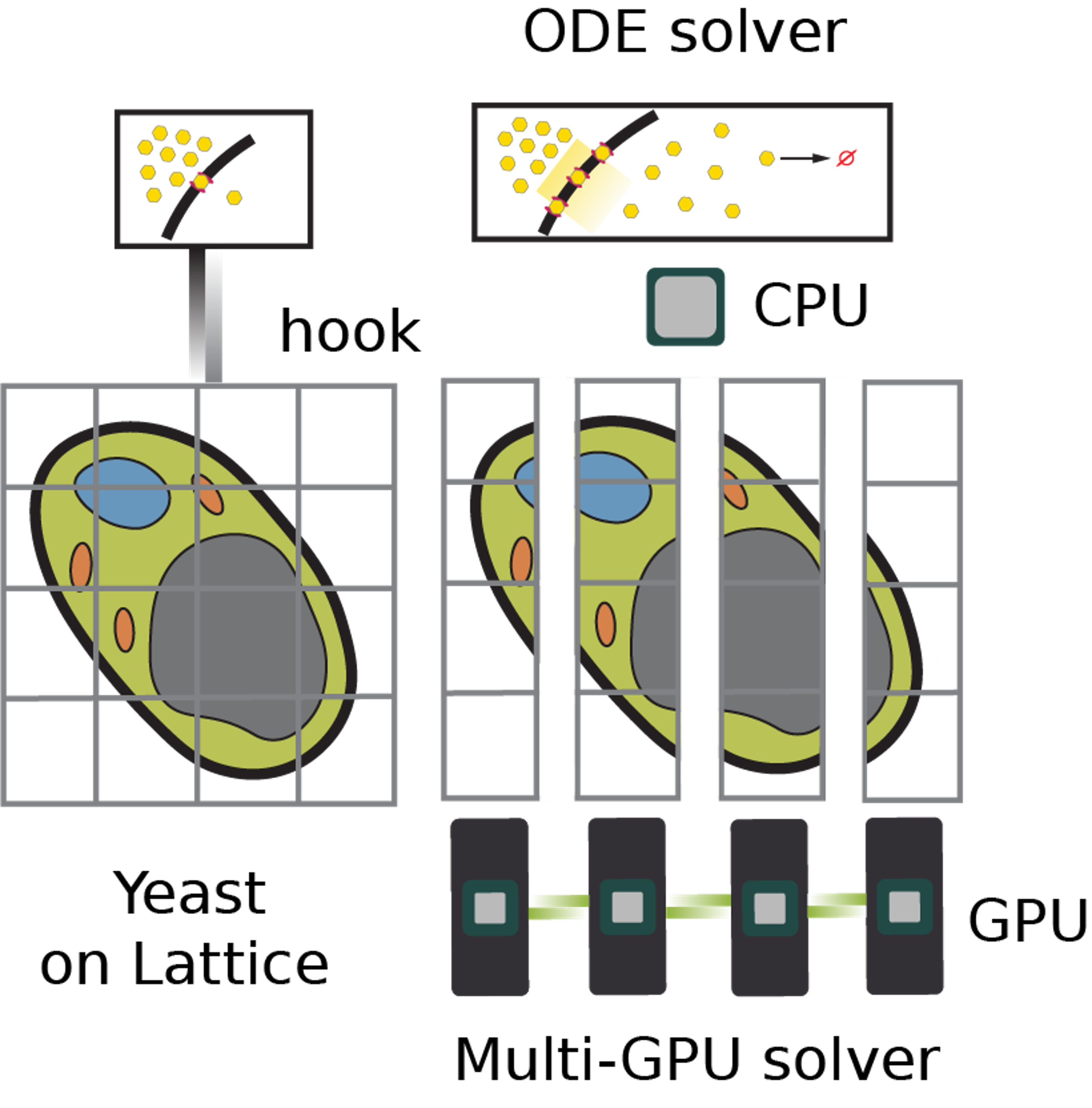 Spatial Heterogeneity Alters the Dynamics of the Yeast Galactose Switch: Insights from 4D RDME–ODE Hybrid SimulationsTianyu Wu, Marie-Christin Spindler, Emmy Earnest, and 4 more authorsbioRxiv, Jan 2025
Spatial Heterogeneity Alters the Dynamics of the Yeast Galactose Switch: Insights from 4D RDME–ODE Hybrid SimulationsTianyu Wu, Marie-Christin Spindler, Emmy Earnest, and 4 more authorsbioRxiv, Jan 2025We present the first 4D simulations of the galactose switch in Saccharomyces cerevisiae using a hybrid framework that integrates reaction diffusion master equations (RDMEs) and ordinary differential equations (ODEs). Using the GPU-based Lattice Microbes program, genetic information processes were simulated stochastically while a simplified metabolism was modeled deterministically. Cell geometry was constructed based on recently acquired cryo-electron tomograms(cryo-ET), which allows us to quantify and differentiate between cytosolic ribosomes and endoplasmic reticulum(ER) associated ribosomes. This allows us to simulate realistic numbers of available ribosomes for ER-associated translation of proteins destined for the cell membrane, like the galactose transporter G2. Our simulations show that an extracellular 11 mM galactose triggers expression of 10k-15k galactose transporters within 60 minutes. We also benchmarked the multi-GPU solver’s performance under various spatial decompositions. Our work underscores the challenges of whole cell modeling of eukaryotic cells and the effects of their inherent spatial heterogeneity.Author summary Cells must quickly adapt when their food source changes. In baker’s yeast, a genetic switch turns on dozens of genes so the cell can use the sugar galactose instead of glucose. We built the first four-dimensional computer model that follows every key molecule in a realistic, tomogram-based yeast cell as this switch is involved. The model combines random, molecule-by-molecule chemistry for rare events with faster, deterministic equations for abundant reactions, and runs on graphics-processing units powerful enough to simulate an entire cell in an hour. By explicitly distinguishing ribosomes that float freely from those anchored on the endoplasmic reticulum, we tracked where the galactose transporter Gal2 is made and how it travels to the cell membrane. The simulations predict that about 10,000–15,000 transporters appear within an hour after the cell senses 11 mM galactose, and reveal that the maze-like endoplasmic reticulum slows their delivery. Our framework shows how cellular geography alters genetic information processing production and paves the way for whole-cell simulations of more complex organisms.Competing Interest StatementThe authors have declared no competing interest.National Science Foundation, https://ror.org/021nxhr62, DBI-2243257European Research Council, 3DCellPhase 760067Cancer Center at Illinois Beckman Institute Postdoctoral Fellows Program