Why we should include spatial information in whole cell modeling
Spatial Heterogeneity Modulates Activation Dynamics of the Yeast Galactose Regulatory Circuit
Project Overview
Spatial organization is a fundamental—but often overlooked—determinant of regulatory behavior in eukaryotic cells. In this project, I build the first spatially resolved, hybrid RDME–ODE model of the Saccharomyces cerevisiae galactose switch, integrating cryogenic electron tomography (cryo-ET)–derived cell geometry, chromosome topology, endoplasmic reticulum (ER) structure, and ribosome distributions into a comprehensive whole-cell simulation framework.
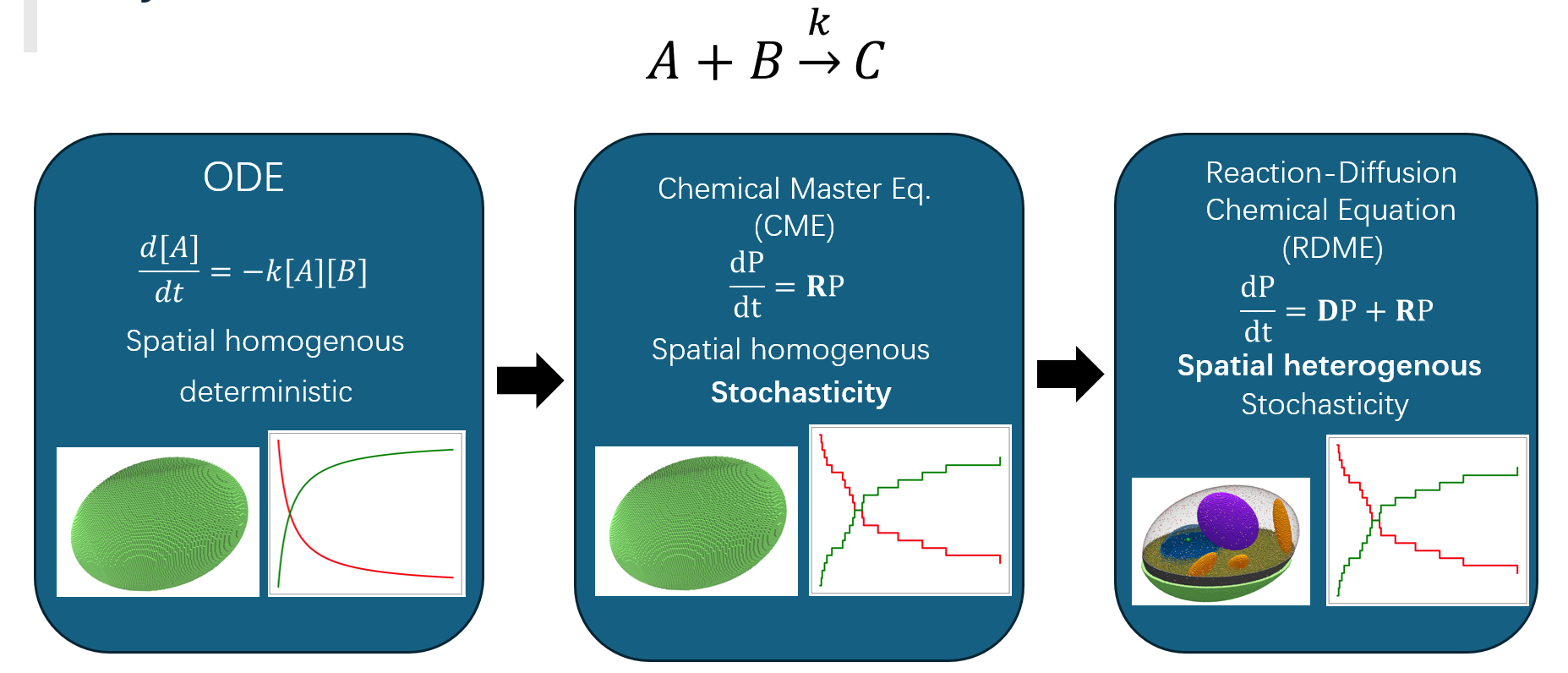
Using Lattice Microbes with a custom multi-GPU reaction–diffusion solver, the model captures how intracellular architecture modulates gene activation, transcriptional output, translation efficiency, and metabolic response. This project bridges structural cell biology and quantitative modeling to demonstrate how 4D spatial–stochastic effects reshape canonical gene regulatory circuits.

Research Goals
Build a spatially faithful yeast cell from cryo-FIB–ET reconstructions, including nucleus, ER subdomains, vacuole, mitochondria, and membrane systems.
Couple stochastic gene regulation (transcription, mRNA dynamics, promoter states, nuclear trafficking) with deterministic metabolism (galactose transport and Leloir pathway initiation).
Determine how spatial constraints, organelle geometry, and ribosome allocation influence switch sensitivity, timing, noise, and protein yields.
Provide a platform for future eukaryotic whole-cell modeling, scalable to larger networks and complex cellular states.
Key Findings
1. Spatial heterogeneity fundamentally alters regulatory dynamics
Introducing realistic geometry and diffusion barriers increases GAL gene activation probability, elevates GAL2 mRNA synthesis, and accelerates early transporter accumulation. Even without adding organelles, spatial structure alone increases Gal2p production and intracellular galactose uptake relative to well-mixed ODE/CME models. The system diverges from the homogeneous model within minutes of induction, demonstrating that spatial effects are not minor corrections but core determinants of system behavior.
2. Chromosome topology has minimal effect on switch output
Embedding all 16 yeast chromosomes into the nucleus creates realistic diffusion barriers and gene localization patterns, yet the system displays near-identical GAL2 activation, transcriptional output, and metabolic response. This suggests that, for this network, regulator abundance and diffusion rates dominate over locus positioning, at least in interphase nuclear architecture.
3. ER geometry suppresses Gal2p synthesis and delays membrane delivery
Explicitly modeling pmaER, cecER, and tubular ER, and restricting translation of Gal2p to ER-bound ribosomes, significantly reduces the fraction of actively translating GAL2 transcripts. Newly synthesized Gal2p accumulates transiently in the ER lumen and reaches the plasma membrane more slowly, demonstrating that protein trafficking pathways impose kinetic bottlenecks that shape gene expression outputs.
4. Ribosome competition creates a major translational bottleneck
Incorporating transcriptome-inferred effective ribosome pools reveals that GAL transcripts occupy only a small fraction of the translational machinery. This reduces Gal2p translation efficiency by nearly half and shifts translation events outward toward peripheral ER. This physical repositioning increases trafficking distances and further limits membrane transporter accumulation. The result: translational resource allocation is a hidden, global regulator of the metabolic switch.